addresses
chat
companies
conversions
custdb
dbBrowser
dialogs
doodle
flatfileDatabase
gadflyDatabase
hopalong
jabberChat
life
minimal
minimalStandalone
noresource
pictureViewer
proof
pysshed
radioclient
redemo
rpn
samples
saveClipboardBitmap
searchexplorer
simpleBrowser
simpleIEBrowser
slideshow
sounds
SourceForgeTracker
spirograph
stockprice
textIndexer
textRouter
tictactoe
turtle
webgrabber
webserver
widgets
worldclock
|
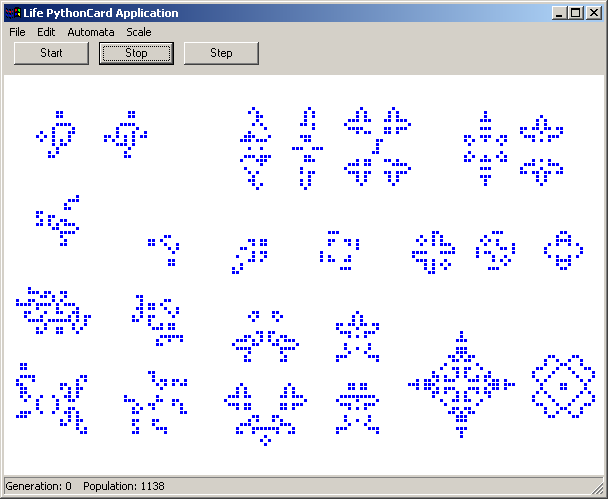
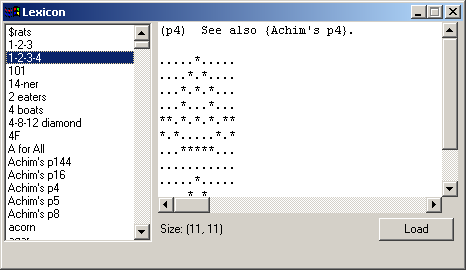
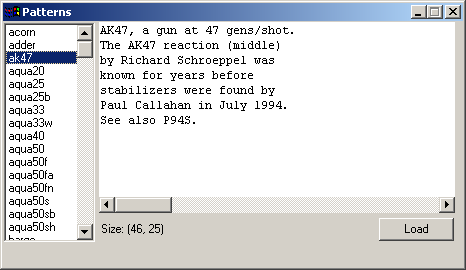
readme.txtA fairly simple implementation of Conway's Game of Life. I hadn't seen one done in Python before and after playing around with the CAGE library decided to see what I could do in a day or two of coding. The universe (grid) size is determined by the window size and the current grid scale. Display slows down as the population size increases, but the size of the grid doesn't impact display speed. You'll probably have to drop the scale down to 1 and use a maximized window in order to display some of the larger Life patterns and even on a fast box and video card you're unlikely to see much more than a generation/frame a second with really large patterns like Breeder. The algorithm I use to calculate the state of the grid at each generation does not wrap. I have not implemented scrolling yet. You can draw in the grid to create your own patterns. I also added clipboard support so that you can paste patterns such as those found in the lifep glossary.doc or anywhere else where you find patterns made up of lines of . and *. On 2002-12-18 I added a Lexicon window which will be displayed if you copy Stephen Silver's lexicon.txt file into the life samples directory before starting up the life sample. There are over 780 definitions and patterns in the lexicon, so I highly recommend you download it. The INTRODUCTION and BIBLIOGRAPHY are listed with the rest of the definitions and patterns. The Lexicon home page is: http://www.argentum.freeserve.co.uk/lex_home.htm And you can download the zip file that contains lexicon.txt directly at: http://www.argentum.freeserve.co.uk/lex_asc.zip For information on Conway's Game of Life and its rules, see Math.com's Life Page. http://www.math.com/students/wonders/life/life.html The original article from the October 1970 issue of Scientific American which introduced the game to the public: http://hensel.lifepatterns.net/october1970.html The fastest and most complete Life program I know of is Life32 by Johan Bontes. http://psoup.math.wisc.edu/Life32.html Life32 like a few other Life programs works on a very large universe size (grid) of 1 million x 1 million, yet is extremely fast because of the clever way the cells in the grid are stored and calculated each generation. I don't know if it uses the same algorithm as Alan Hensel's Java applet, but that applet is also very fast. http://hensel.lifepatterns.net/ The algorithm, source, etc. is available at: http://hensel.lifepatterns.net/lifeapplet.html I expect that if something similar was done in Python then my program would be fast too. I'll leave it as an exercise for the reader |